Glutamate Receptors
There are three main types of glutamate receptors that are distinguished on the basis of their different pharmacology (Table 1). They all respond to the neurotransmitter but have different responses to artificial agonists or antagonists.
Table 1 Glutamate Receptors
| Receptor | Agonist | Antagonist | Number of Genes |
|---|---|---|---|
| AMPA Receptors | AMPA | CNQX | 4 |
| NMDA Receptors | NMDA | APV | 9 |
| Kainate Receptors | Kainic acid | 5 |
The two main subtypes, the AMPA receptors and NMDA receptors, have quite distinct functional properties (Table 2).
Table 2 Comparison of AMPA and NMDA Receptors
| AMPA Receptors |
|---|
| main form of GluR at most central excitatory synapses |
| fast kinetics - fast onset, offset and desensitization in response to glutamate |
| small single channel conductance |
| most native receptors contain the GRIA2 subunit which limits Ca2+ permeability |
| NMDA Receptors |
|---|
| slow kinetics |
| large single channel conductance |
| high Ca2+ permeability |
| blocked by Mg2+ ions |
| glycine is a co-agonist |
Structure of Glutamate Receptors
The mammalian glutamate receptors share a similar topology and quaternary structure. The individual subunits have three membrane spanning domains and the channel is a tetramer, assembled from four subunits (Figure 1). There is no homology between the amino acid sequence of glutamate receptors (GluRs) and Cys-loop receptors.
A protein homologous to the mammalian glutamate receptor is found in prokaryotes where it is involved in chemoreception. The evolution of this protein provides some insight into the glutamate receptor structure (Figure 1). The prokaryotic receptor appears to have arisen from the fusion of a periplasmic binding protein (PBP) and a two-membrane spanning domain K+ channel. Periplasmic binding proteins are involved in high affinity active transport of amino acids and other nutrients in bacteria. The topology of the membrane spanning domain in the fused protein is inverted in comparison to K+ channels. This receptor retains its K+ ion selectivity and is activated by amino acids. The receptor assembles as a tetramer, which further supports the idea that it is derived from a K+ channel. The bacterial periplasmic binding proteins have a clamshell-like structure that clamps around the ligand when it enters the binding site, a function that is retained in the mammalian receptor.
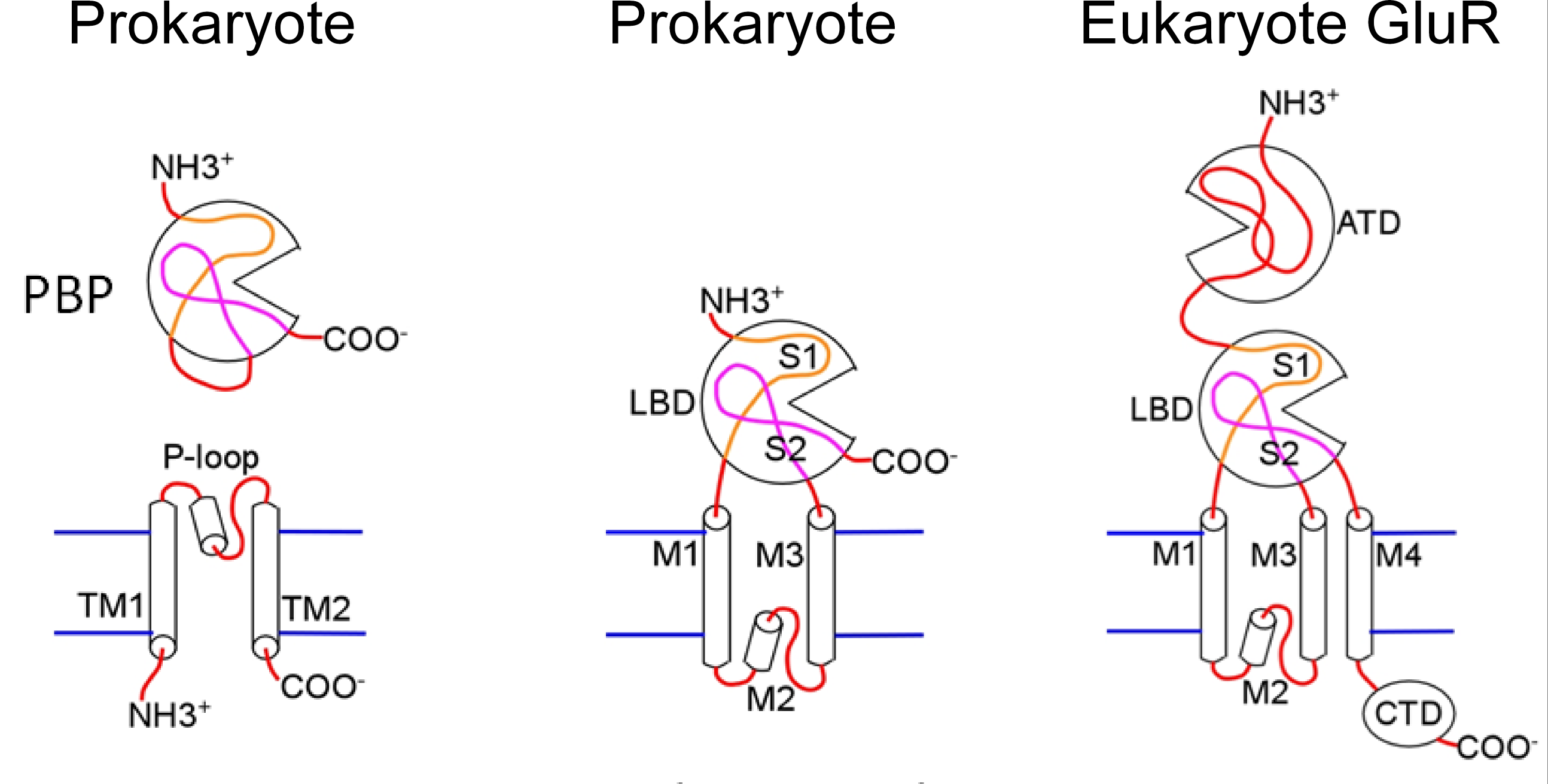
Figure 1 Bacterial periplasmic binding protein (PBP) and K+ channel proteins appear to be the precursors of the prokaryote receptor. Abbreviations: ligand binding domain (LBD), amino terminal domain (ATD).
A second fusion event involving the leucine–isoleucine–valine binding protein (LIVBP), another bacterial periplasmic binding protein, and the proto-receptor appears to have added the amino terminal domain (ATD) found in eukaryote glutamate receptors. A third membrane spanning domain was gained in the eukaryote receptor and the K+ selectivity of the pore was lost.
The glutamate receptor is a good example of the modular nature of protein design. Evolution has used a cut-and paste process to create a new protein, combining protein domains from several different sources to generate a protein with unique functional properties.
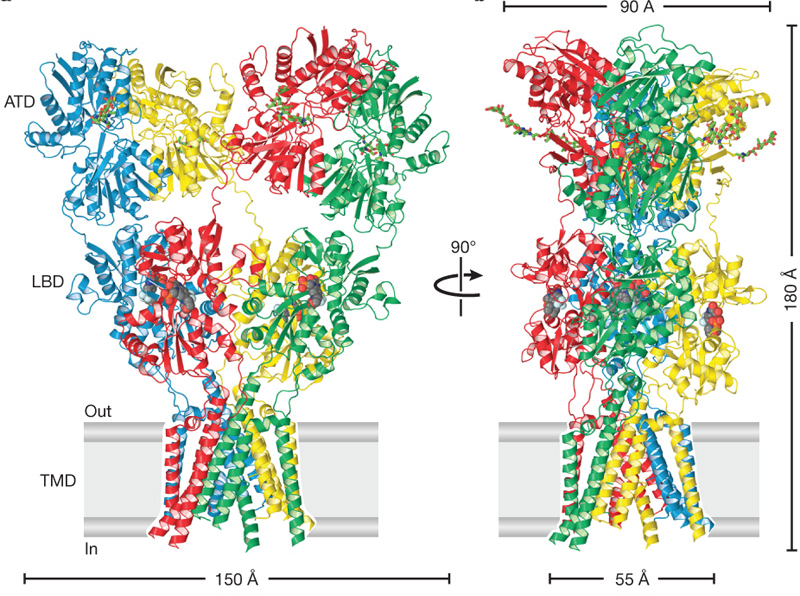
Figure 2 Structure of the AMPA receptor. Competitive antagonist molecules lodged within each LBD clamshell are shown in space-filling representation. Abbreviations: ligand binding domain (LBD), amino terminal domain (ATD), transmembrane domain (TMD).
The amino terminal domain (ATD) and ligand binding domain (LBD) have 2-fold symmetry whereas the transmembrane domain (TMD) retains the 4-fold symmetry of the original K+ channel.