Transporters
All cells must transport a diverse set of biochemical molecules, including ions, amino acids, and sugars, across the cell membrane. There are three classes of transporters: uniporters, symporters, and antiporters (Figure 1).
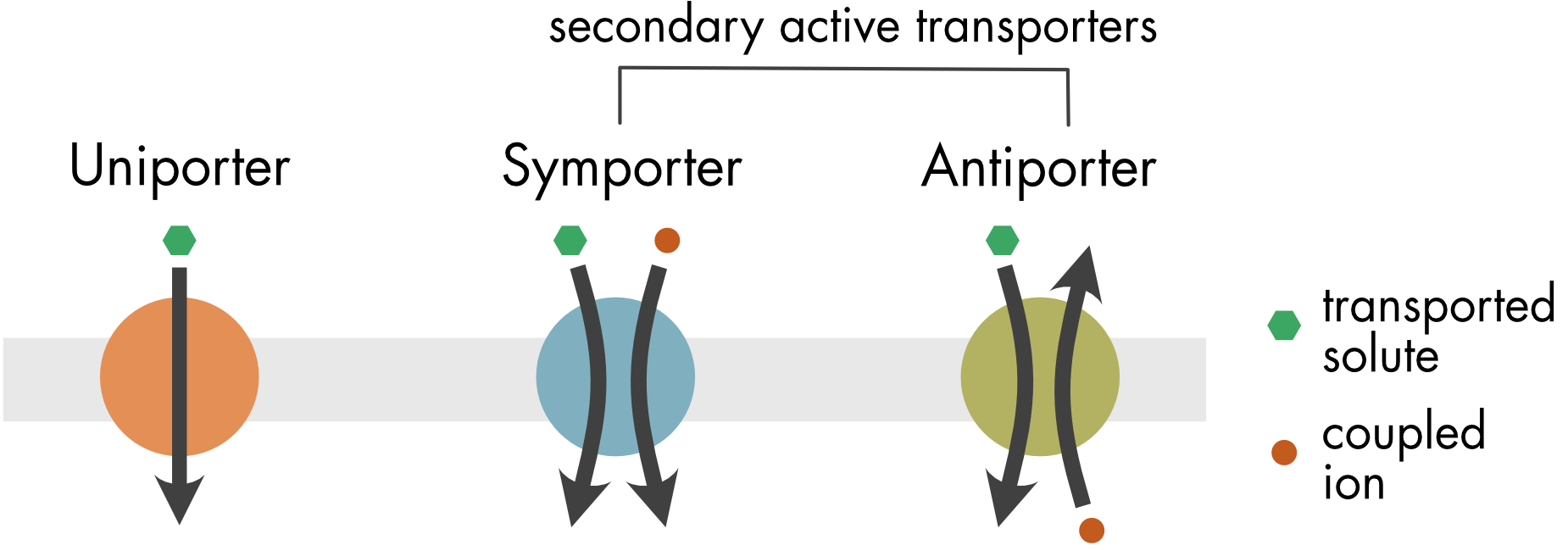
Figure 1 Uniporters transport a single substrate down a concentration gradient. Symporters transport a solute and a coupling ion in the same direction across the membrane. Antiporters sequentially transport two different molecules in opposite directions across the membrane. For both symporters and antiporters, the transported solute moves up its concentration gradient and the coupled ions move down their concentration gradient.
Uniporters are the simplest type of transporter. These membrane proteins transport solutes down their concentration gradients. They do not require any input of energy. This process is known as facilitated diffusion—the transporter facilitates the diffusion of the solute across the membrane.
There is an important kinetic difference between simple and facilitated diffusion (Figure 2). Facilitated diffusion will saturate as the solute concentration increases due to the dependence on a finite set of transporter proteins. Saturation follows Michaelis-Menten kinetics, similar to saturation of an enzyme activity. In contrast, simple diffusion will be a linear function of solute concentration.
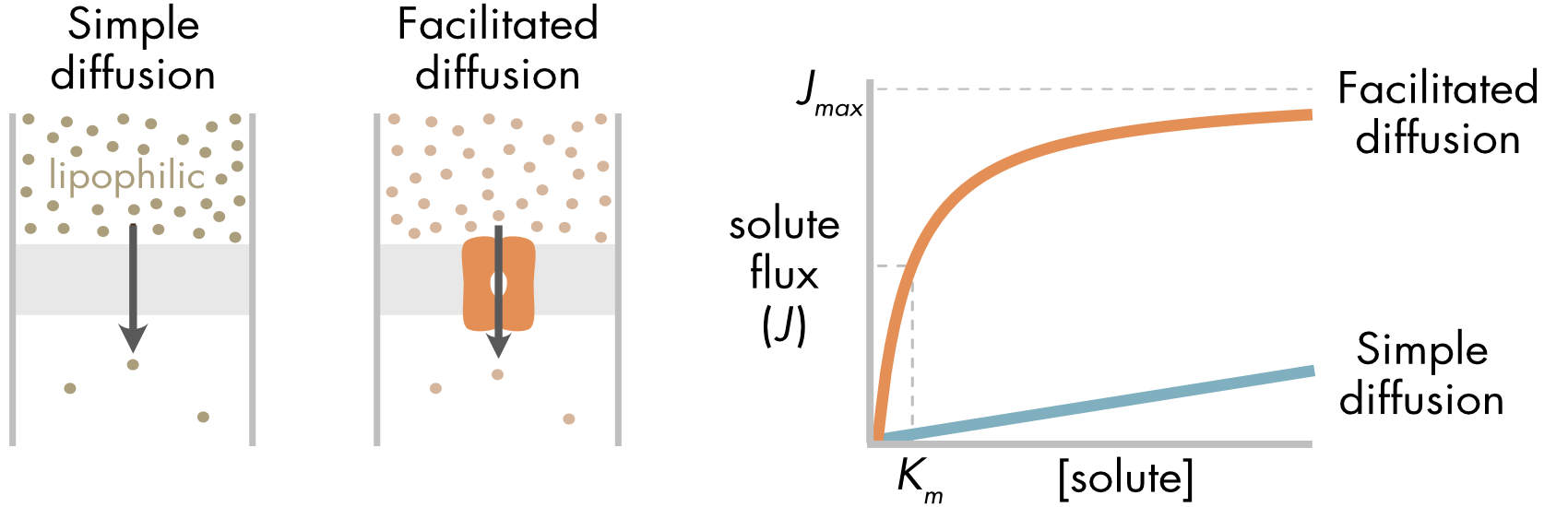
Figure 2 (left panel) Diffusion of a lipophilic solute directly through the membrane and facilitated diffusion mediated by a passive transporter. (right panel) Facilitated diffusion saturates as solute concentration increases. In contrast, passive diffusion increases linearly with solute concentration. The maximum flux (Jmax) is the maximum rate of solute flux supported by the finite number of passive transporters in the cell membrane.
All transporters show saturation kinetics. The rate of transport depends directly on the number of transporters in the cell membrane, although it can also be regulated by cell signaling pathways.
Secondary active transporters use the energy stored in the electrochemical gradients of ions to move a solute up its concentration gradient. They are called secondary because they use chemical energy stored in the form of an ion gradient rather than directly using ATP. They are called active transporters because they move solutes up a concentration gradient.
A typical transporter moves a solute across the membrane by an ‘alternate access’ mechanism (Figure 3). In the open-out state of the transporter the transported solute can only access the solute binding site from one side of the membrane. The transporter then undergoes a conformational change to the open-in state, where the binding site becomes accessible from the cytoplasm and inaccessible from the outside of the cell. Flipping between these two states enables the movement of the substrate across the membrane. The conformational changes are known as a rocker-switch mechanism. The two, roughly symmetrical, halves of the protein flip in a coordinated way from one conformation to the other.
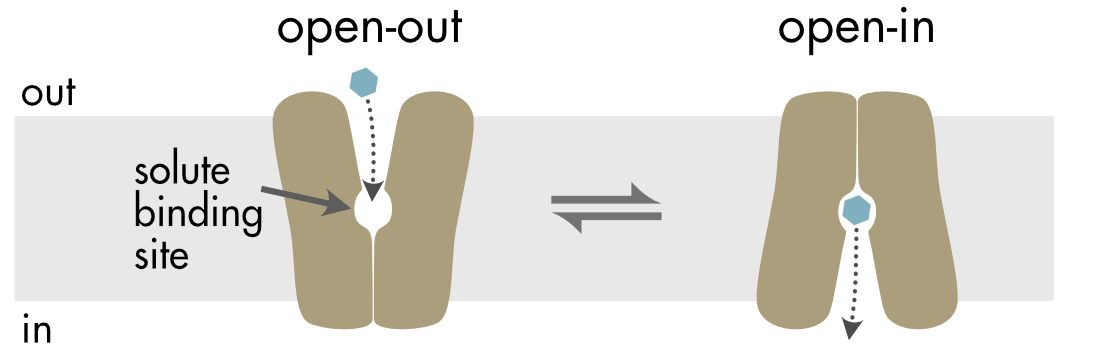
Figure 3 In the alternate access model the transporter flips between two states (open-out and open-in). In the open-out state the solute binding site is accessible from the extracellular side of the membrane and in the open-in state the binding site is accessible from the cytoplasm. The movement of the two halves of the transporter resembles the movement of a rocker switch.
The transition between the two states is triggered by thermal energy as well as binding energy from solute and ion binding to specific sites within the transporter. Transitions can occur in the absence of solute binding due solely to the contribution of thermal energy. This is necessary to allow the transporter to return to the open-out state in the absence of solute binding. Typically, the rate of transition between the states is increased by solute binding.
Both uniporters and secondary active transporters are reversible. In the case of the uniporter the direction of transport is simple to understand, there is a net movement of solute down its concentration gradient. For active transporters, the direction of transport is affected by the electrochemical potential of one or more coupled ions in addition to the concentration of the transported solute (see Chapter 3 for details).
This basic picture of membrane transporters conceals the considerable complexity of this family of membrane proteins.
Molecular Biology of Transporters
Uniporters and secondary active transporters belong to the SLC (SoLute Carrier) superfamily, which in humans has more than 400 members subdivided into 65 gene families (SLC1-65), see Figure 4 and Transporter Table. The SLC superfamily is the second largest family of membrane proteins, after the G protein-coupled receptor superfamily. The large size of this gene family reflects the enormous diversity of molecules that transporters have to move. There are transporters for sugars, amino acids, nucleotides, inorganic ions, urea, fatty acids, iodide, and organic cations and anions. To maintain ligand specificity a wide variety of receptors are required i.e. one size does not fit all. In general, members of each different SLC family transport similar substrates. The large majority of transporters within this superfamily are either symporters or antiporters. Uniporters are relatively rare.

Figure 4 Phylogeny of the SLC superfamily. (Figure provided by G. Gyimesi, and M.A. Hediger, Univ. Bern., unpublished).
The SLC superfamily has considerably more structural diversity than other membrane protein superfamilies such as G protein–coupled receptors, voltage-gated ion channels, and tyrosine kinase receptors. In part this reflects the fact that most of the different transporter gene families identified in the human genome are evolutionary ancient and predate the divergence of bilaterians.
The SLC family can be divided into several clans based on protein structure. These include: MFS (Major Facilitator Superfamily), APC (Amino acid-Polyamine-organoCation), CPA/AT (Cation Proton Antiporter/Anion Transporter), and DMT (Drug/Metabolite Transporter). The MFS clan is the largest group of phylogenetically related SLCs, containing approximately one-third of all SLCs. Different families of transporters can have from 3 to 14 transmembrane domains. For two families the gene products can only generate functional transporters as heteromeric partners, where one partner has a single transmembrane domain. The different clans are not only structurally distinct but can have distinctly different mechanisms of solute transport.
SLC family transporters have important roles in the central nervous system in addition to their support of basic cell metabolism and physiology. These functions include movement of nutrients across the blood–brain barrier, reuptake of neurotransmitters after transmitter release at synapses, filling of synaptic vesicles, regulation of the chloride gradients that underlie inhibitory synaptic transmission, and regulation of cytoplasmic calcium ion concentrations.
Structure and Function of Transporters
Because of the fundamental differences between the structure and function of different SLC families, it is necessary to focus on particular families or clans within SLC superfamily when considering their detailed structure and function. This section focuses on the passive glucose transporter, which is a member of the MFS clan. This transporter combines the rocker switch mechanism described above with a gated pore mechanism.
MFS Transporters
The SLC2 family of transporters is an important family within the MFS clan (Transporter Table). The SLC2 family contains 14 related transporters (GLUT1-14). All but one are uniporters that facilitate the movement of monosaccharides (glucose and/or fructose).
The ubiquitous passive glucose transporters (Figure 5) are members of this family. This transporter is found in the membrane of most cells. Passive glucose transporters typically move glucose from the extracellular fluid into the cell, where the concentration of glucose is lower due to the constant utilization of glucose by the cell’s metabolic processes. The transporter is reversible and will transport glucose out of the cell if the gradient is reversed, as occurs in hepatocytes during fasting. The brain has no local energy stores, and the glucose transporter is critical for maintenance of the energy supply to neurons.
Figure 5 The glucose transporter mediates the facilitated diffusion of glucose down its concentration gradient, usually from the extracellular fluid into cells. The transporter cycles between four distinct states: open-out (o-out), occluded-out (occ-out), occluded-in (occ-in), and open-in (o-in). The glucose binding site of the transporter is first exposed to the outside of the cell (o-out). Solute binding causes the outer gate to close (occ-out). This is followed by a rocker-switch movement (occ-in). Then the inner gate opens (o-in), and the glucose is released to the cytoplasm. Critically, both gates are never open simultaneously. The blue hexagons represent glucose molecules. Step through the animation by clicking the ‘step’ button or run the complete animation by clicking the ‘run’ button.
Like channels, uniporters create a protein wrapper that allows polar or charged molecules to diffuse across the membrane while avoiding interaction with the lipid bilayer. A key difference between transporters and ion channels is the rate of transport. When a channel opens ion flow through the channel is limited primarily by the rate of diffusion of the ions to and through the channel. In contrast, the relatively slow cycling of the transporter between the different states required for transport of a single molecule significantly limits the rate of solute movement (Figure 5). The limited transport rates of individual transporters means that these transport systems can quickly become saturated if the solute concentration rises. To create greater solute flow requires insertion of more transporters into the membrane. The insulin-dependent upregulation of glucose transporters in some cell types after a meal is an example of this form of regulation.
GLUT5 Structure and Function
GLUT5 is a fructose transporter and a member of the SLC2 family of transporters. Its structure and function is relatively well understood (Figure 6). Like other members of the SLC2 family of transporters, it has 12 membrane spanning domains with intracellular N- and C-termini. The transmembrane domains are organized into two bundles, each containing six transmembrane helices, that are separated by an intracellular domain comprised of several smaller helices.
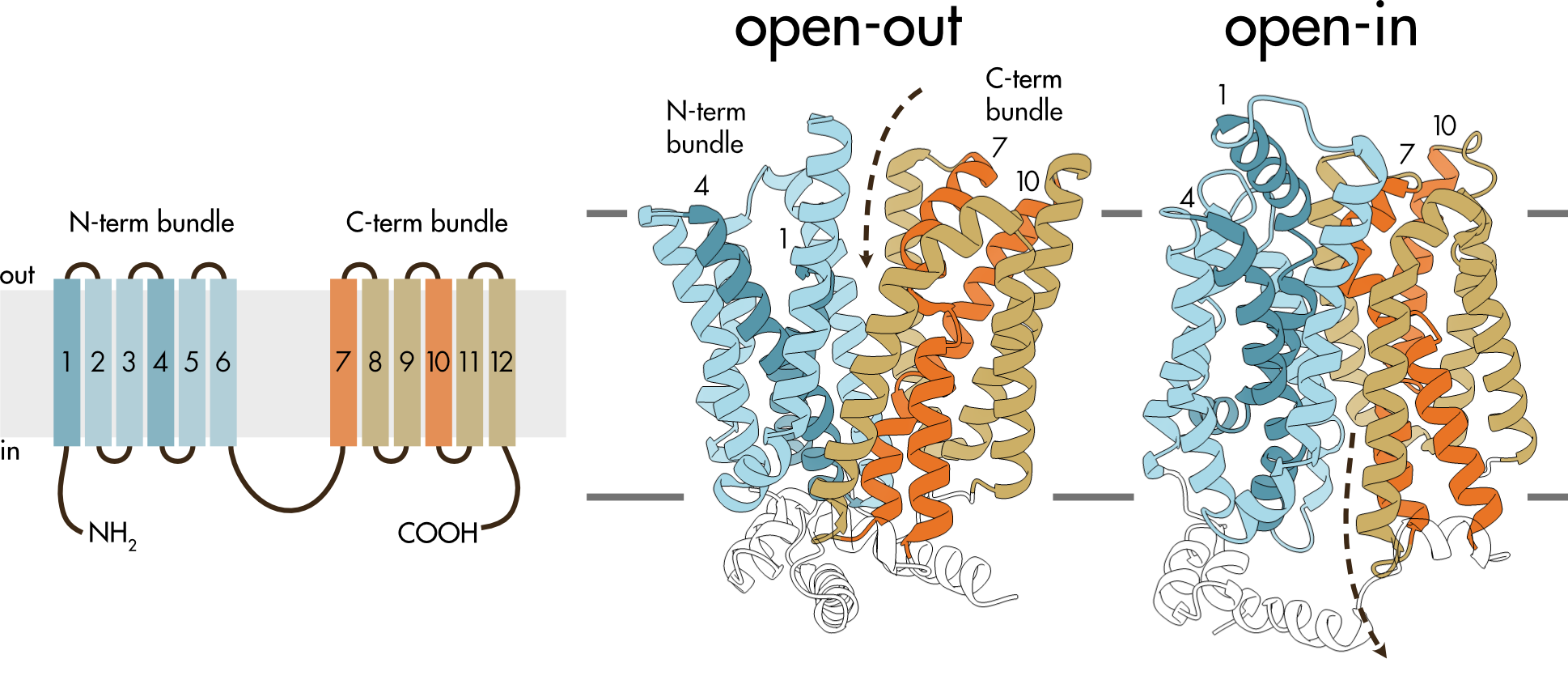
Figure 6 (left panel) Membrane topology of SLC2 transporters. There are 12 transmembrane helices (TM1-12) and both the amino- and carboxy-termini are located intracellularly. The large intracellular domain between helices 6 and 7 divides the structure into two halves, the N-terminal bundle and the C-terminal bundle. (right panel) Structure of the open-in and open-out conformations of GLUT5. TM1 and TM4 are colored dark blue and TM7 and TM10 are colored orange. The dashed arrows show the typical direction of fructose movement from the extracellular solution to the binding site and from this site into the cytoplasm.
Solute transport involves a coordinated movement of the two bundles. In the open-out conformation the two bundles are arranged to allow access to the central binding site from the extracellular solution. The open-in conformation allows access to the binding site from the intracellular solution. The rocker-switch movement required to move between these two states is relatively easy to imagine by comparing the open-out and open-in structures (Figure 6).
In addition to the rocker-switch movement there are smaller gating movements by several transmembrane helices that line the pore, particularly TM7 and TM10 in the C-terminal bundle but also TM1 and TM4 in the N-terminal bundle (Figure 7).
Figure 7 Movement of the GLUT5 helices involved in pore gating. The transition from the open-out (o-out) to the occluded-out (occ-out) state is mediated primarily by movement of the upper half of TM7 induced by fructose binding. Transition to the occluded-in (occ-in) state requires coordinated rigid body movement of the N-terminal and C-terminal bundles, the rocker-switch movement. Transition to the open-in (o-in) state involves movement of the lower half of TM10. The upper half of TM7 and the lower half of TM10 are the primary contributors to gating but movement in the TM1 and TM4 in the N-terminal bundle also contribute. Critically, both gates are never open simultaneously. The red pentagon represents the fructose molecule. Step through the animation by clicking the ‘step’ button or run the complete animation by clicking the ‘run’ button.
Diversity of Transporters
The transporter family is large and structurally diverse. Although all transporters appear to conform to the alternating access model, where they alternate between outward- and inward-facing conformations, how they move between these two conformations varies considerably. Three basic mechanisms are rocker switch (described above), rocking-bundle, and elevator mechanisms.
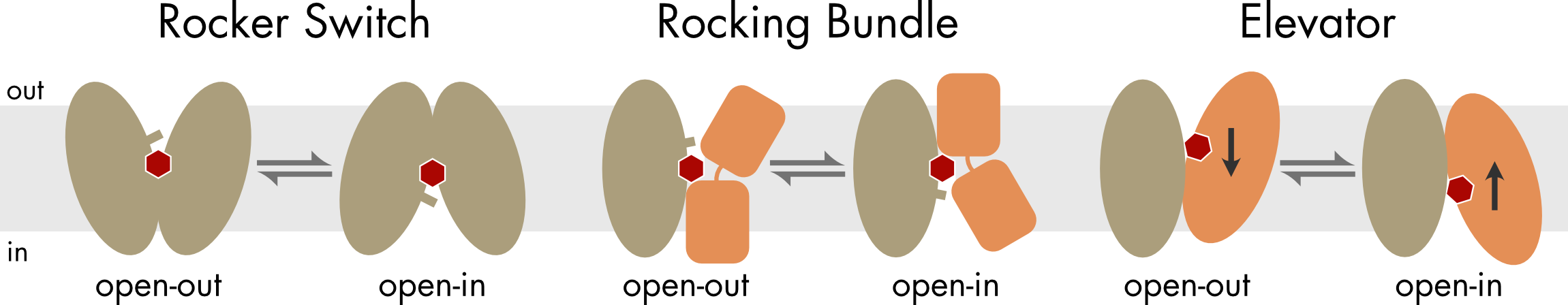
Figure 8 For the rocker switch mechanism the two domains of the transporter are pseudo-symmetric and large rigid body movements by both domains accompany the transition between the two main states. In the other two mechanisms the two domains are structurally distinct, with one domain remaining relatively fixed and the other domain moving significantly during the transition between the open-out and open-in states. For the elevator mechanism the substrate binding site is formed solely by the moving domain, which moves vertically within the membrane. In addition to the large rigid body movements there can be more subtle gating movements induced by substrate binding (indicated by bars).
For the rocker switch and rocking bundle mechanisms the solute binding site is located roughly in the center of the interface between the two domains, which rearrange around that central site during transitions between the two main states. For the elevator mechanism the solute binding site is located on the moving domain. During the transition between the open-out and open-in states the binding site moves vertically relative to the plane of the membrane.